| oligos_5-8nt_m1_shift10 (oligos_5-8nt_m1) |
 |
|
|
|
|
|
|
|
|
|
|
|
|
; oligos_5-8nt_m1; m=0 (reference); ncol1=11; shift=10; ncol=21; ----------atACACGtaww
; Alignment reference
a 0 0 0 0 0 0 0 0 0 0 47 39 159 1 144 0 0 32 70 43 51
c 0 0 0 0 0 0 0 0 0 0 39 37 1 143 2 162 0 26 26 32 40
g 0 0 0 0 0 0 0 0 0 0 38 40 0 0 12 0 162 19 30 32 26
t 0 0 0 0 0 0 0 0 0 0 38 46 2 18 4 0 0 85 36 55 45
|
| ANAC55_ANAC55_ArabidopsisPBM_shift11 (ANAC55:ANAC55:ArabidopsisPBM) |
 |
0.870 |
0.791 |
5.365 |
0.927 |
0.947 |
6 |
2 |
41 |
15 |
16 |
16.000 |
3 |
; oligos_5-8nt_m1 versus ANAC55_ANAC55_ArabidopsisPBM (ANAC55:ANAC55:ArabidopsisPBM); m=3/149; ncol2=10; w=10; offset=1; strand=D; shift=11; score= 16; -----------wACACGTAAy
; cor=0.870; Ncor=0.791; logoDP=5.365; NsEucl=0.927; NSW=0.947; rcor=6; rNcor=2; rlogoDP=41; rNsEucl=15; rNSW=16; rank_mean=16.000; match_rank=3
a 0 0 0 0 0 0 0 0 0 0 0 36 80 2 97 2 1 1 88 87 9
c 0 0 0 0 0 0 0 0 0 0 0 19 3 77 2 96 2 12 8 1 40
g 0 0 0 0 0 0 0 0 0 0 0 15 4 2 0 1 95 13 0 0 10
t 0 0 0 0 0 0 0 0 0 0 0 30 13 19 1 1 2 74 4 12 41
|
| ABI5_M0261_1.02_CISBP_shift10 (ABI5:M0261_1.02:CISBP) |
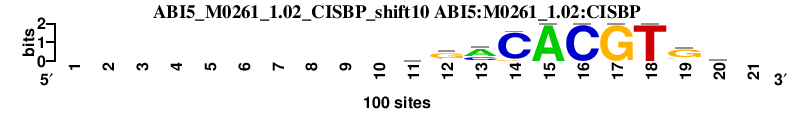 |
0.848 |
0.771 |
6.449 |
0.921 |
0.938 |
15 |
4 |
15 |
25 |
33 |
18.400 |
4 |
; oligos_5-8nt_m1 versus ABI5_M0261_1.02_CISBP (ABI5:M0261_1.02:CISBP); m=4/149; ncol2=10; w=10; offset=0; strand=D; shift=10; score= 18.4; ----------bgmCACGTGk-
; cor=0.848; Ncor=0.771; logoDP=6.449; NsEucl=0.921; NSW=0.938; rcor=15; rNcor=4; rlogoDP=15; rNsEucl=25; rNSW=33; rank_mean=18.400; match_rank=4
a 0 0 0 0 0 0 0 0 0 0 18 9 61 0 100 0 0 0 15 16 0
c 0 0 0 0 0 0 0 0 0 0 26 9 31 92 0 100 0 0 7 16 0
g 0 0 0 0 0 0 0 0 0 0 25 65 0 8 0 0 100 0 71 34 0
t 0 0 0 0 0 0 0 0 0 0 31 17 8 0 0 0 0 100 7 34 0
|
| ABF1_M0257_1.02_CISBP_shift10 (ABF1:M0257_1.02:CISBP) |
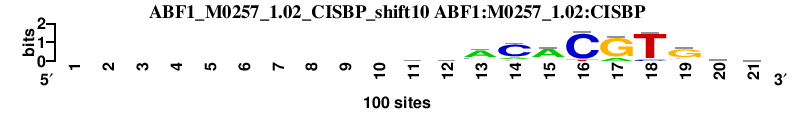 |
0.845 |
0.845 |
3.652 |
0.935 |
0.954 |
20 |
1 |
90 |
7 |
8 |
25.200 |
6 |
; oligos_5-8nt_m1 versus ABF1_M0257_1.02_CISBP (ABF1:M0257_1.02:CISBP); m=6/149; ncol2=11; w=11; offset=0; strand=D; shift=10; score= 25.2; ----------gdACACGTGkb
; cor=0.845; Ncor=0.845; logoDP=3.652; NsEucl=0.935; NSW=0.954; rcor=20; rNcor=1; rlogoDP=90; rNsEucl=7; rNSW=8; rank_mean=25.200; match_rank=6
a 0 0 0 0 0 0 0 0 0 0 21 26 68 16 73 0 18 0 12 16 18
c 0 0 0 0 0 0 0 0 0 0 21 16 18 75 8 91 0 10 4 19 28
g 0 0 0 0 0 0 0 0 0 0 37 33 7 0 10 0 82 0 71 40 28
t 0 0 0 0 0 0 0 0 0 0 21 25 7 9 9 9 0 90 13 25 26
|
| AT4G18890_MA1333.1_JASPAR_rc_shift10 (AT4G18890:MA1333.1:JASPAR_rc) |
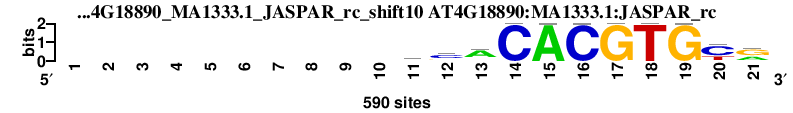 |
0.773 |
0.773 |
0.077 |
0.905 |
0.901 |
77 |
3 |
146 |
86 |
96 |
81.600 |
|
; oligos_5-8nt_m1 versus AT4G18890_MA1333.1_JASPAR_rc (AT4G18890:MA1333.1:JASPAR_rc); m=93/149; ncol2=11; w=11; offset=0; strand=R; shift=10; score= 81.6; ----------ycACACGTGyr
; cor=0.773; Ncor=0.773; logoDP=0.077; NsEucl=0.905; NSW=0.901; rcor=77; rNcor=3; rlogoDP=146; rNsEucl=86; rNSW=96; rank_mean=81.600; match_rank=
a 0 0 0 0 0 0 0 0 0 0 81 52 405 0 589 0 0 0 1 0 225
c 0 0 0 0 0 0 0 0 0 0 179 338 60 590 0 589 0 0 0 383 43
g 0 0 0 0 0 0 0 0 0 0 91 140 74 0 0 0 590 0 589 28 319
t 0 0 0 0 0 0 0 0 0 0 239 60 51 0 1 1 0 590 0 179 3
|




